The CBC has the mission of transforming the conduct of biomedical research and education in the Chicago area in order to promote and enable scientific research of the highest order. It is grounded in the belief that the combined intellectual power of the researchers, educators, and staff of the member institutions must be harnessed to fulfill a vision for Chicago to become a national and international leader in the biomedical sciences.
To fulfill its mission, the CBC organizes a variety of educational programs and events, including the annual symposia, has assisted in faculty recruitment to the CBC universities, provides support for numerous cutting-edge and unique research facilities at the three CBC universities, and offers several competitive award programs. The current and past CBC Award Programs are described below.
Ongoing Programs
Accelerator Awards I Director’s Fund Awards
Archived Programs and Past Funding Opportunities
Affinity Group Awards I Catalyst Awards I CBCAN I COVID-19 Response Awards I HTS Awards I Postdoctoral Awards I Scholars Program I Exploratory Workshops I Lever Awards I Spark Awards
Ongoing Programs
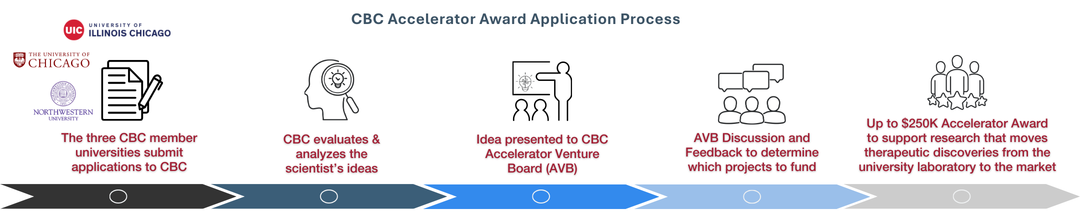
Meet Our Recent Accelerator Awardees
Meet Our Recent Director’s Fund Awardees
Archived Programs and Past Funding Opportunities
- NEUROSCIENCE AFFINITY GROUP (2023)
- Build a community of Chicago nueroscientists
- Develop research collaborations between Chicago neuroscientists and incorporate orthogonal expertise
- Articulate fundamental neuroscience questions to drive collaborations across Chicago scientists
- Develop a novel, team science approach to advance attractive scientific projects
- Accelerate high-risk/high-reward research
- PIs: Rui Gao (UIC) and Kaiwen Kam (RFUM) for the project:
Connectomic reconstruction of a hindbrain neural circuit controlling breathing across the lifespan - PIs: Sarah Lutz (UIC) and Brian Popko (NU) for the project:
Blood-brain barrier dysfunction in vascular dementia - PIs: Ruth Anne Eatock (UChicago) and Anna Lysakowski (UIC) for the project:
Visualizing evoked activity in vestibular inner ear and afferent projections to brain
The overarching goal of the Neuroscience Affinity Group was to provide a framework for selecting high-risk/high-reward research projects in the Neuroscience space.
The objectives of this program were to:
Targeting Aberrant Immune Responses in Patients with Severe COVID-19
Award Number: CR-001
Date Awarded: September 2020
Amount Awarded: $ 500,000.00
Abstract: The public health and societal impacts of COVID-19 are all attributable to patients with severe disease. We have collected samples from the lungs of 86 patients with severe COVID-19 and 252 patients with severe pneumonia from other causes (likely the world’s largest) and autopsy tissue from 36 Chicagoans who died from or underwent lung transplant for COVID-19. Our detailed genomic analysis of these samples suggests why COVID-19 is more severe in some, with therapeutic implications. We will test this in human lung tissues infected with SARS-CoV-2 at Argonne labs and will credential a therapy to promote lung healing after COVID- 19.
Novel Strategies for Enhancing Vaccine Efficacy Against SARS-CoV-2
Award Number: CR-002
Date Awarded: September 2020
Amount Awarded: $ 500,000.00
Abstract: The rapid spread of COVID19 highlights the necessity of vaccines against its causative coronavirus, SARS- CoV-2. We will explore strategies to understand and improve neutralizing antibody responses against the viral Spike protein, which mediates viral entry into target cells. We will develop novel functional nanomaterials to target the vaccine to specific cells that regulate immune response, specifically dendritic cells to induce immunity and lymphatic endothelial cells to induce enhanced memory. We will employ advanced microscopy to gain better understanding of how specific cell types affect the generation and breadth of the response.
Covalent Inhibitors of the Nsp16 2′-O-Methyltransferase of SARS-CoV-2
Award Number: CR-003
Date Awarded: September 2020
Amount Awarded: $ 498,750.00
Abstract: There are limited treatments for COVID-19, the global pandemic respiratory disease caused by the virus SARS-CoV-2. A potentially effective treatment would be a drug against the virus 2’- O-methyltransferase nsp16/nsp10 protein complex. This protein made by the virus is required to modify the ends of critical virus nucleotide sequences that code for the building blocks necessary to assemble a complete virus. This modification also helps the virus hide from our immune system. We propose to design effective inhibitors of this complex. The goal is to generate a lead compound for advanced drug design and for efficacy in SARS-CoV-2 infection models.
High Throughput Screening (HTS) Supplemental Grant Program ran from 2013 until November 2016. The Program aimed to help fund discovery of innovative small-molecule probes and “hits”. The intent of this program was to support pilot projects, which are focused on biomedically-relevant targets and which would be conducted at a HTS core facility located at one of the CBC universities. The CBC HTS Award would match half of the justified expenses (up to $20,000) of a small molecule screening project, via payment to the specified HTS facility.
| Scholar | Class of | Home University | Graduation Date | Scholar | Class of | Home University | Graduation Date | |
| Aggarwal,Chaitanya | 2012-2013 | UIC | 2014 (Dec) | Antanasijevic, Aleksandar | 2014-2015 | UIC | 2016 (May) | |
| Arslan, Sevim Yildiz | 2011-2012 | UIC | 2013 (May) | Baumgarten, Sarah | 2013-2014 | UIC | 2017 (Jun) | |
| Boland, Michelle (Beaton) | 2013-2014 | UChicago | 2016 (Jun) | Best, Timothy | 2012-2013 | UChicago | 2012 (Apr) | |
| Cassidy, Justin | 2010-2011 | NU | 2013 (Aug) | Chaudhuri, Rima | 2010 | UIC | 2010 (Dec) | |
| Chow, Christina | 2011-2012 | UIC | 2013 (Jun) | Cone, Jackson | 2012-2013 | UIC | 2014 (Dec) | |
| Curtis, Matthew | 2010-2011 | UIC | 2011 (May) | Daniel, Weston | 2010 | NU | 2010 (Dec) | |
| Dore, Lou | 2011-2012 | NU | 2012 (Jun) | Gebhardt, Michael | 2014-2015 | UChicago | 2017 (Mar) | |
| Glotfelty, Lila (Gollogly) | 2011-2012 | UIC | 2014 | Hammer, Lauren (Smith) | 2014-2015 | NU | 2016 (May) | |
| Hlavaty, Kelan | 2013-2014 | NU | 2015 (Jun) | Kathman, Stefan | 2014-2015 | NU | 2016 (Nov) | |
| Khaja, Fatima A. | 2013-2014 | UIC | 2014 (Mar) | Klosterman, Susan | 2012-2013 | UIC | 2013 (May) | |
| Leech, Kristan | 2013-2014 | NU | 2015 (May) | Mao, Qiyan | 2011-2012 | UChicago | 2013 (Jun) | |
| Mayo, Leah | 2014-2015 | UChicago | 2015 (Apr) | Mendoza, Adelita | 2014-2015 | NU | 2017 (Dec) | |
| Nalle, Sam | 2010-2011 | NUChicago | 2011 (Sep) | Pah, Adam | 2011-2012 | NU | 2013 (Jun) | |
| Payne, James T. | 2013-2014 | UChicago | 2015 (Aug) | Ramu, Haripriya | 2010-2011 | UIC | 2011 (Jun) | |
| Rayahin, Jamie E. | 2014-2015 | UIC | 2017 (Dec) | Restrepo, Nicolas Pelaez | 2011-2012 | NU | 2016 (Mar) | |
| Rowell, Joanna | 2012-2013 | UChicago | 2013 (Jun) | Scarpelli, Andrew | 2012-2013 | NU | 2015 (Jul) | |
| Serebryannyy, Leonid A. | 2014-2015 | UIC | 2016 (Aug) | Shepard, Jaclyn | 2010-2011 | NU | 2012 (Jun) | |
| Shy, Brian R. | 2013-2014 | UIC | 2017 (Jun) | Singer, Bryan F. | 2011-2012 | UChicago | 2012 (Nov) | |
| Sun, Fei | 2011-2012 | UChicago | 2012 (Jun) | Sundaram (White), Erin | 2010-2011 | UChicago | 2013 (Feb) | |
| Tarasewicz, Elizabeth | 2012-2013 | NU | 2015 | Tietjen, Gregory | 2012-2013 | UChicago | 2013 (Jul) | |
| Wells, Daniel | 2013-2014 | NU | 2015 (Dec) | Werner, Michael S. | 2013-2014 | UChicago | 2016 (Mar) | |
| Yap, Jonathan Woon Teck | 2012-2013 | NU | 2016 | Zimmerman, John | 2014-2015 | UChicago | 2016 (Aug) |
The program ran from 2011 until April 2016. The intent of the CBC Exploratory Workshops was to bring together small groups of faculty members who were interested in making interdisciplinary and inter-institutional connections around cutting-edge research areas.
February 07, 2016
CBC Exploratory Workshop:
Seeing better together – Strategic Cross-Institutional Microscopy Initiatives
June 10, 2014
The CBC Vascular Biology Exploratory Workshop
May 15, 2013
The CBC Exploratory Workshop on Lipoproteins
January 30, 2013
The CBC Exploratory Workshop on Cellular Heterogeneity
January 12, 2013
The CBC Ovarian Cancer Workshop
September 27, 2012
Control of cellular differentiation and gene expression by 5- hydroxymethlcytosine (5-hmc)
PIs: Vadim Backman (NU), Lucy Godley (UChicago) and Jack Kaplan (UIC) for project:
Chicago Center for Physical Science-Oncology Innovation and Translation
Amount Awarded: $ 1,487,992.00
Award Description: The CBC’s sixth Lever Award provides support for the Chicago Center for Physical Science-Oncology Innovation and Translation. Principal Investigators on the Lever are Vadim Backman (NU), Lucy Godley (UChicago) and Jack Kaplan (UIC). The $1.5 million CBC Lever was awarded in conjunction with a $10 million U54 grant from the National Cancer Institute to fund the Chicago Region Physical Science-Oncology Center (CR-PSOC). The goal of CR-PSOC is to advance the understanding of carcinogenesis by examining the role of physical and chemical forces involved in the transformation of a normal cell into a cancerous one. Specifically, the studies will focus on interrogating changes in both the epigenome and the metallome (the metal ion content of the cell) that contribute to the development of cancer.The CR-PSOC, led by Thomas V. O’Halloran and Jonathan D. Licht, is composed of a multi-disciplinary team of 12 physical scientists and 8 cancer researchers from fields encompassing physics, chemistry, biomedical engineering, biophysics, biochemistry, pharmacology, and hematology-oncology from the three CBC institutions as well as experts in the physical sciences and chromatin fields from outside Chicago, namely MIT, Memorial-Sloan Kettering Cancer Center, and the University of Massachusetts Medical School.
Designed around the theme of “Spatio-Temporal Organization of Chromatin and Information Transfer in Cancer,” the CR-PSOC consists of three interrelated project areas, each focused on different aspects of chromatin structure and function, plus two core facilities, and pilot project, education and outreach programs.
The CBC Lever funds will support the Center’s new instrumentation and shared resources:
- Nanocytometry Core (Northwestern)
Leader: Vadim Backman
It will include Partial Wave Spectroscopy (PWS) and Stochastic Optical Reconstruction Microscopy (STORM) for high resolution microscopy - PDX Core (Northwestern)
Leader: Andrew Mazar- Patient-derived xenografts will provide meaningful models of human cancer and enable translation of PSOC innovations
- Includes funding for investigator pilot studies using PDX models
- New high precision methylation analysis capabilities through acquisition of an Illumina NextSeq500, to be used in the University of Chicago’s Genomics Core
Leader: Lucy Godley - New ultra-sensitive IC-ICP-mass spectrometer capability to be used in the Quantitative Bioelement Imaging Center at Northwestern
Leader: Thomas O’Halloran
PIs: Anthony Kossiakoff and Geoffrey Greene (UChicago), Brian Kay (UIC), and Jason Brickner (NU) for project:
Center for Production of Affinity Reagents for Human Transcription Factors: Chicago Synthetic Antibody Pipeline
Amount Awarded: $ 2,321,520.00
Award Description: The CBC has awarded the fifth Lever Award to scientists from the three CBC member institutions: Tony Kossiakoff (UChicago), Brian Kay (UIC), Geoffrey Greene (UChicago) and Jason Brickner (NU) for a proposal, Center for Production of Affinity Reagents for Human Transcription Factors: Chicago Synthetic Antibody Pipeline (CSAP). The $2.3M CBC Lever Award was awarded in conjunction with an NIH U54 grant, Recombinant Antibody Network (RAN). The goal of the $12.2M NIH grant is to generate renewable, high quality affinity reagents against all human transcription factors. The Lever Award will use the NIH RAN infrastructure to generate affinity reagents for the CBC community against a variety of targets including membrane and soluble proteins, protein complexes and functional RNA. Synthetic antibodies will be generated by phage display and will be stored as plasmids, allowing for low cost regeneration. The consortium is in its early stages so please check the CBC webpage for updates.See also:
Chicago Synthetic Antibody Pipeline (CSAP)
PIs: Andrey Rzhetsky (UChicago), Edwin Cook (UIC), and Richard Morimoto (NU) for project:
Silvio O. Conte Center on the Computational Systems Genomics of Psychiatric Disorders
Amount Awarded: $ 1,999,431.00
Award Description: The CBC has awarded the fourth Lever Award to a group of scientists from the three CBC member institutions: Andrey Rzhetsky (UChicago), Edwin Cook (UIC), and Richard Morimoto (NU) to support the Silvio O. Conte Center on the Computational Systems Genomics of Psychiatric Disorders. The CBC Lever of $2 million matches a $11.75 million grant from the National Institute of Mental Health: the Silvio O. Conte Center for Basic and Translational Mental Health Research (P-50) award. The new Conte Center, a multi-institutional effort, will be based at the University of Chicago, and will apply data mining strategies to study mental disorders. Specifically the center aims to design and validate a battery of novel analytical tools for the inference of causal relationships among human genomic variations, environmental factors, and more than one neurodevelopmental phenotype, explicitly exploiting the genetic and environmental non-independence of complex (multigenic) disorders.The P50 has four core projects that will study the genome structure of molecular networks involved in psychiatric disorders as well as the effect of genetic and non-genetic variation on the topology and characteristics of these networks.
- Core Project 1: Pharmacogenomics and Modeling the Joint Effects of Genes and Environment in Psychiatric Disorders
- Core Project 2: Modeling the temporal succession of phenotypes and environmental cues in the context of latent genetics
- Core Project 3: Modeling disease risk in the context of molecular networks and gene association or linkage data
- Core Project 4: Deciphering brain phenotypes through integrative modeling of multiple data types
The CBC Lever Awards are matching grants made to inter-institutional groups that are submitting large-scale grant proposals. The CBC Lever funds supporting the Conte Center will be specifically used for:
- Enhancing the computational and experimental resources related to the Silvio O. Conte Center for Basic and Translational Mental Health Research (P-50) award, making them available to Chicago-area researchers, whether they are included as Principal Investigators on the P-50 or not.
- Purchasing computer infrastructure for text mining of large-scale molecular networks, for large-scale study of variants in human genes associated with clinical phenotypes, for analysis of genetic and pharmagenomic data in the context of molecular networks, and to make these analyses maximally useful to a wide group of researchers.
- Providing technical support and experimental supplies for P-50 and non-P-50 investigators – using the tri-institutional cloud computing infrastructure – for accesssing deep sequencing and proteomics services, as well as testing theories from computational analysis in model biological systems.
- Supporting extensive educational and outreach activities involving the Chicago-area research community.
For more information about the center visit the Conte Center website.
PIs: Chad Mirkin and Milan Mrksich (NU), David Eddington (UIC) and Joel Collier (UChicago) for project:
Nanomaterials for Cancer Diagnostics and Therapeutics
Amount Awarded: $ 2,117,241.00
Award Description: The CBC has awarded a Lever of $2.1 million over three years to establish two core facilities that will develop, fabricate, and disseminate standardized and well-defined matrices and substrates for culturing cancer cells. The facilities will be housed at the University of Illinois at Chicago and at the University of Chicago and operated by a team of dedicated technicians at both sites. The CBC Lever Award matches a $12 million award over five years from the National Cancer Institute (NCI) to help establish a collaborative network of Centers of Cancer Nanotechnology Excellence (CCNEs).CBC Lever Awards are matching grants made to inter-institutional groups that are submitting large-scale grant proposals. The Principal Investigators on the Nanomaterials for Cancer Diagnostics and Therapeutics CBC Lever Award are Chad Mirkin (NU), David Eddington (UIC), Milan Mrksich, (NU) and Joel Collier (UChicago).
The core facilities will have a two-part objective:
(1) Fabricate and Disseminate Tools – the two cores primary effort will be directed towards the preparation of the patterned substrates, peptide amphiphiles and culture devices.
(2) Technical Instruction – the two cores will provide applications specialists who will travel to local laboratories and instruct laboratory members in the use of the tools.
Both the culture tools and technical support will be provided to interested laboratories at no cost. Thus the CBC will provide a mechanism and funding for the translation of NCI-funded research into broad use.
The following foundries are offered to the CBC community through the Nanomaterials for Cancer Diagnostics and Therapeutics Lever Award:
- Microenvironmental Control Foundry
- The Nanopatterning Foundry (to be open in Summer 2012)
- Matrix Synthesis Foundry (to be open in Summer 2012)
PIs: Sergey Kozmin (UChicago), Karl Scheidt (NU) and Jie Liang (UIC) for project:
Chicago Tri-Institutional Center of Excellence in Chemical Methodologies & Library Development
Amount Awarded: $ 2,000,000.00
Award Description: The CBC has awarded the second Lever Award to a group of scientists from the three CBC member institutions; Sergey Kozmin (UChicago), Karl Scheidt (Northwestern), Hisashi Yamamoto (UChicago), Vladimir Gevorgyan (UIC), Viresh Rawal (UChicago), Milan Mrksich (NU; at UChicago at the time of the award), and Jie Liang (UIC). The $2 million Lever award matches a $9.2 million award from the National Institutes of Health (NIH) to support the establishment and operation of the Chicago Tri-Institutional Center for Chemical Methods and Library Development (CTCMLD). The CTCMLD will provide significant new resources for biomedical research in the Chicago community and boost drug discovery efforts in the area by advancing high-throughput organic synthesis and integrating the production of new small-molecule libraries with broad biological screening.In order to achieve the maximum impact, the CTCMLD will feature a high degree of collaboration and synergy with several research programs at the University of Chicago, Northwestern University, and University of Illinois at Chicago. Sergey Kozmin (UChicago) will serve as the director of the Center during the initial five year period. The CTCMLD collaborative group will also include exceptional strength in organic synthesis, represented by Karl Scheidt (Northwestern), Hisashi Yamamoto (UChicago), Vladimir Gevorgyan (UIC) and Viresh Rawal (UChicago), as well as the leading expertise in surface engineering of Milan Mrksich (NU; at UChicago at the time of the award) and cheminformatics of Jie Liang (UIC).
CBC Lever Awards are matching grants made to inter-institutional groups that are submitting large-scale grant proposals. Lever Awards are primarily used to establish transformative infrastructure that can be made broadly available to the Chicago scientific community. The CTCMLD represents a high-impact tri-institutional collaboration with projects to be headed by PIs from all three CBC member institutions. The CTCMLD Lever Award will support the following three key initiatives:
1. Enhancing the capabilities of the High-Throughput Synthesis component of the Core Facility of CTCMLD at University of Chicago, by establishing a professionally staffed facility with the state-of-the-art equipment for high-throughput synthesis, purification and analytical characterization of newly generated chemical libraries.
2. Establishing the Hit-to-Lead Development Resource of the CTCMLD at Northwestern University, which will provide unique support for innovative research at the interface of chemistry and biology in Chicago area.
3. Developing a Computational Cheminformatics Core at UIC, which will be used to guide production of small-molecule libraries with favorable physicochemical properties and will facilitate analysis of the compound screening data.
Three cores are available to the CBC research community through the Chicago Tri-Institutional Center for Chemical Methods and Library Development (CTCMLD):
- Library Production Core (located on the University of Chicago campus)
- Hit-to-Lead Development Resource (HLR) (located at Northwestern University)
- Computational Cheminformatics Core (located at the University of Illinois at Chicago)
For more information on the cores click here or go directly to the CTCMLD website.
PIs: Kevin White, PhD (UChicago); Robert Grossman, PhD (UChicago; at UIC at the time of the award); Richard Morimoto, PhD (NU); Luis Amaral, PhD (NU) for project:
Chicago Center for Systems Biology
Amount Awarded: $ 3,000,000.00
Award Description: The CBC has awarded $3 million over three years to help establish the Chicago Center for Systems Biology (CCSB). The CBC Lever Award matches a $15 million award from The National Institute of General Medical Sciences. The CCSB will be one of 10 National Centers for Systems Biology — the first of its kind in Illinois and an outstanding new research resource for the Chicago region. Systems Biology is an emerging field, focusing on the study of complex interactions in biological systems, including everything from the smallest molecules to complete organisms.Kevin White at the University of Chicago will direct the Chicago Center for Systems Biology, which revolves around collaborations among Chicago-area experts in genomics, developmental biology, evolutionary biology, stress and physiology, chemistry and physics and computational professionals who specialize in network modeling and high-performance computing. Combining experimental and computational tools, the CCSB will study the dynamic behavior of gene networks in cells, tissues, and organisms, paying specific attention to transcriptional networks, clusters of master genes that regulate the activity of other genes by directly turning them on or off.
CBC Lever Awards are matching grants made to inter-institutional groups that are submitting large-scale grant proposals. The Principal Investigators on the Lever Award to the CCSB are Luis Amaral (Northwestern), Robert Grossman (University of Chicago; at UIC at the time of the award), Richard Morimoto (Northwestern), and Kevin White (University of Chicago). Lever Awards are primarily used to establish transformative infrastructure that can be made broadly available to the Chicago scientific community. The CCSB Lever Award will support the following four key initiatives:
1. Developing an enhanced imaging core, that uniquely combines microfluidics and confocal microscopy for live imaging of model organisms, tissues, and cells.
2. Cultivating a recombineering and high-throughput cloning core to support genetic modifications to chromosomal sections for human, mouse, Drosophila, and C. elegans genomes.
3. Advancing a computational core that will integrate intimately with the biological driver projects and which will produce software modules that will be useful to a much broader community.
4. Establishing a “CBC Research Fellows Program in Systems Biology” to train the next generation of young scientists in the art of interdisciplinary research in Systems Biology.
The following cores are available to the CBC research community at the Chicago Center for Systems Biology (CCSB):
1. Advanced Imaging Core (AIC)
For more information about the Chicago Center for Systems Biology (CCSB) click here.
PIs: Nissim Hay (UIC), Joseph Bass (NU), Graeme Bell and Louis Philipson (UChicago) for project:
Leptin peptide in diabetes: from mechanism to therapeutics
Amount Awarded: $ 399,597.00
Abstract: Leptin receptor levels are highest in the brain, which is consistent with the primary action of leptin to decrease food intake. However, we hypothesize that, under normal physiological conditions, leptin also plays a key role in the regulation of blood glucose levels through direct effects on the liver and regulation of hepatic glucose production. This effect is more readily evident in insulin-deficient states, common in both type 1 and late type 2 diabetes. We further propose that insulin modulates leptin signaling as abrogation of insulin signaling increases leptin levels in blood and leptin receptor expression in liver. Although this hypothesis challenges the commonly accepted view that leptin exerts its effects through it action in various regions of the brain, there are other observations that provide support for our hypothesis: 1. Liver-specific insulin receptor knockout mice display a 10-fold increase in serum leptin levels as well as a 35- fold increase in leptin receptor levels mRNA in the liver; 2. The level of hepatic leptin receptor is dramatically increased in mice with liver-specific knockout of the downstream insulin receptor signaling molecules IRS1 and PI3 kinase; 3. The expression of hepatic leptin receptor in Akt-deficient mice is markedly increased (data not shown); and 4. Leptin treatment in human patients with lipodystrophy and in multi-tissue Akt-mutant mice (both of which have low leptin levels) normalizes blood glucose levels. Thus, impaired hepatic insulin/PI3K/Akt signaling in the liver is coupled to a marked elevation of leptin receptor levels concomitant with an increase in the levels of circulating leptin. Moreover, deficiency of leptin due to impaired development and/or function of adipose tissue is linked to hepatic overproduction of glucose, increasing insulin resistance and thereby exacerbating diabetes, and fasting hyperglycemia in particular. Surprisingly, the significance of these observations implicating leptin signaling in the regulation of hepatic function are not fully appreciated and need to be further explored to impact clinical practice. Thus, a major goal of this proposal is to challenge the current dogma that leptin exerts its effect on glucose homeostasis exclusively or largely though its effect on the brain. Specifically we will generate hepatic leptin receptor KO mice. These mice will be treated to induce type 1 diabetes and will be subjected to leptin therapy. These mice will also be crossed with Akita mice and the compound mice will be subjected to leptin therapy.
PIs: Anthony Kossiakoff (UChicago), Vladimir Gelfand and Charles Clevenger (NU) for project:
Delivery of synthetic antibodies to probe cell dynamics in live cells
Amount Awarded: $ 400,000.00
Abstract: Goals: We propose to develop a technology platform for the next generation of affinity reagents and imaging probes that will substantially extend capabilities for observing dynamic events in live cells. Transitions between distinct protein conformations or the formation of multi-protein complexes are fundamental to cellular structures, signaling and regulation. However, current antibody and fluorescent-protein tags are generally insensitive to these subtle, but functionally critical changes in target molecules. To overcome this deficiency we have developed a new technology platform that both generates highly specific affinity-binders that have the capability to identify specific protein complexes and transient conformations and to deliver these probes to living cells with experimental ease and minimal perturbation so they can interrogate cellular dynamics in their native cell environments. As a challenging test-bed to develop and test our new technology, we will generate unique affinity reagents and imaging probes to study core aspects of signaling and motor function essential in cytoskeletal dynamics that cannot be investigated using current methods. This pilot project is only the beginning for what we believe is the full potential for our technology platform. Our long term objective is to assemble a high-throughput pipeline to produce high-affinity synthetic affinity reagents to high-impact protein and RNA targets for use in imaging, localization and inhibition studies in their native cellular environments. We believe it is well within the capability of the technology to automate most of the steps of the pipeline with the aim to create a powerful, compact and affordable core facility-like platform to provide all Chicago area researchers with designer antibodies that can be tailored to particular systems and needs.
PIs: Thomas O’Halloran and Vinayak Dravid (NU) and Jonathan Silverstein (UChicago) for project:
Support for An Innovative CryoSTEM for Element Specific Imaging of Cells and Tissue
Amount Awarded: $ 379,341.00
Abstract: Support from the two-year Spark Award will be used to fund two scientists who will aid CBC faculty members in using a high-resolution cryo-capable scanning transmission electron microscope (Cryo STEM), soon to be acquired. The microscope is capable of following changes in subcellular distributions of essential or toxic metal ions at the level of a few hundred atoms at a time and offers the unprecedented ability to produce quantitative maps of each relevant element within a biological sample. One CBC-funded scientist will support use of the instrument and train and advise CBC users in sample preparation and data acquisition. A second CBC-funded scientist will make the resulting multidimensional data usable by developing computational methods for 2-D and 3-D volumetric rendering representing the spatial variations in elemental concentrations.This spark award leverages core funding that the team, along with Teresa Woodruff of Northwestern, received from the W. M. Keck Foundation for the purchase and installation of the Cryo STEM. The acquisition of this custom-designed high-resolution electron microscope will bring a novel imaging resource to the Chicago area. While the device will be located on the Northwestern Evanston campus, the full-time staff supported by the CBC will open up use of this unique and exciting technology to researchers from the other CBC member institutions.
PIs: Martin Kreitman and Ilya Ruvinsky (UChicago) and Richard Morimoto (NU) for project:
The Role of Natural Variation and Proteostasis in Complex Disease Traits
Amount Awarded: $ 400,000.00
Abstract: Our project “The Role of Natural Variation and Proteostasis in Complex Disease Traits”, will attempt to create a new paradigm for studying human diseases with multifactorial genetic and environmental causes. Modern molecular genetics has triumphed over the past decade in the discovery of major mutations causing important human genetic diseases. But many diseases, such as adult-onset diabetes and autism, are not caused by mutations in single genes, but instead are characterized by a complex, and yet unknown, interaction between genetic variations in large number of genes and subtle environmental factors. For these diseases, molecular genetics has been much less successful in deciphering their genetic underpinnings. Our project, a collaboration between two evolutionary geneticists, Dr. Martin Kreitman and Ilya Ruvinsky (Department of Ecology and Evolution, U. Chicago), and a molecular geneticist, Dr. Richard Morimoto (Biochemistry, Molecular and Cell Biology, Northwestern U), proposes a novel way forward by investigating natural variation for susceptibility and severity of complex human disease, recreated in two model organisms, the fruitfly Drosophila melanogaster and the worm Caenorhabditis elegans. The two human diseases we will investigate — neurodegeneration and neonatal diabetes — share a common attribute in that they both result from the inability of targeted cells to respond to physiological stresses imposed by the expression of unstable mutant proteins. The inability of cells to respond to these stresses results in programmed cell death, the direct cause of disease. According to our working hypothesis, a complex diffuse web of interacting naturally occurring polymorphisms in fly, worm and human sets an individual’s ability to respond to genetic or environmental challenges, determining susceptibility to and severity of disease. We will, for the first time, investigate naturally occurring variation affecting disease models in powerful model genetic systems, which we believe has the potential to revolutionize the use of disease these models. The goal of our experiments will be to identify cellular and genetic mechanisms in the worm and fly that influence the severity of these model diseases, taking advantage of the many power genetic and molecular tools available in these model organisms. We believe that commonalities in the worm and fly will also prove to be shared with the human form of the diseases, and that discoveries in these model organisms will therefore be relevant to developing novel therapies to disease. Our findings about effects of genetic variation on the ability of a cell to balance protein synthesis, folding, transport, and degradation (proteostasis), may have broader relevance to other sporadic diseases, in addition to diabetes and neurodegenerative disease. More generally, by integrating the study of natural genetic variation in model organisms with human genetic diseases recreated in these organisms, our research holds the promise of introducing powerful new strategy for deciphering the genetic basis of complex human disease.
PIs: Erik Sontheimer (NU), Alexander Mankin (UIC) and Jonathan Staley (UChicago) for project:
Noncoding RNA Structure, Function, and Evolution
Amount Awarded: $ 400,000.00
Abstract: Organisms depend on the proper and dynamic functioning of their genes, many of which encode proteins that carry out essential cellular tasks. Additionally, species depend on the evolution of genes to adapt and survive over time. Because inappropriate gene expression causes or contributes to many diseases, including cancer, the expression of genes into proteins must be controlled properly to maintain human health. Genes are comprised of DNA, and the first step of gene expression involves the production of an RNA copy of the gene’s DNA sequence. In many cases the RNA is simply an intermediate that serves as a template for the production of a protein. In other instances, however, the RNA has its own biochemical function beyond the temporary transmission of genetic information, and such RNAs are known as noncoding RNAs (ncRNAs). RNA has increasingly been found to be as important as proteins in executing and regulating gene expression, and the number of known ncRNAs has skyrocketed. However, the boundaries of the ncRNA universe are not yet known, and the functions of most ncRNAs remain mysterious. To meet the challenges and exploit the opportunities of this exciting time in RNA research, we will capitalize on our existing strengths in this area by establishing a Center of Excellence for the investigation of the structure, function and evolution of ncRNAs. Specifically, we aim to discover new modes of ncRNA function, define novel ncRNA populations, explore their structures and interactions with other cellular components, and determine how ncRNA genes evolve. To catalyze these long-term goals, we will expand our capabilities in two areas central to ncRNA research: custom chemical synthesis and bioinformatics. Because ncRNAs are now known to impinge upon nearly all areas of biology, the establishment of an ncRNA research center will benefit the Chicago biomedical community as a whole.
PIs: Keith Thulborn and Y. Jeong (UIC) and Thomas Meade (NU) for project:
Metabolic MR Imaging for Studies of Human Brain Disease
Amount Awarded: $ 400,000.00
Abstract: The treatment for human disease at the earliest stages requires sensitive, quantitative, and non-destructive diagnostic monitoring methods. Magnetic resonance imaging (MRI) is one of the few technologies capable of localized, nondestructive metabolic characterization without ionizing radiation. The high spatial and temporal resolution of MR imaging provides the means to investigate physiology at the systems level (i.e., whole organisms).This project develops a new dimension of MR imaging to monitor metabolic changes expressed in the earliest stages of disease and during response to treatment. It will focus on obtaining insights into the interrelated problems of developmental and molecular biology and clinical diseases by i. generating MR probes that function as real-time in vivo physiological reporters (exogenous), ii. develop quantitative parameters of metabolic concentrations and rates that reflect tissue health (endogenous) and iii. correlate in vivo image analysis by pharmacokinetic methods. Although applicable to any location in the body, the human brain will be the initial target organ.
Bioscales will be created using MR signals. The signals will be derived from metabolites occurring naturally in the body, or from bioactivated probes that detect specific biochemical processes. New classes of probes will be synthesized and optimized and human and non-human primate MRI studies will be performed at the enhanced sensitivity of 9.4 Tesla to calibrate these bioscales for metabolic modeling of the healthy brain.
These metabolic models will be combined with enhanced MR imaging applicable to clinical field strengths of 3.0 Telsa. This translational approach will result in metabolic imaging for monitoring the earliest stages of diseases, thereby stimulating the development of earlier, and therefore less expensive, interventions. The strategic plan of the NIH emphasizes healthcare cost containment through earlier treatment of disease. Metabolic imaging using both exogenous and endogenous agents is an essential technology for developing such early intervention strategies for humans.
PIs: Robert Goldman and Jonathan Widom (NU), Stephen Kron, Harindar Sighn and Elizabeth McNally (UChicago) for project:
The Chicago Laminome Project
Amount Awarded: $ 400,000.00
Abstract: The nuclear lamins are members of one of the largest families of proteins. Lamins form mesh-like networks within the nucleus, providing a molecular interface, the lamina, between the membranes surrounding the nucleus and the chromosomes contained inside. In recent years the lamins have been shown to be major components of an extensive regulatory network involved in a wide range of functions, including the control of components of an extensive regulatory network involved in a wide range of functions, including the control of nuclear architecture, the organization, positioning and structure of chromosomes, gene expression, and DNA replication and repair. However, the molecular mechanisms underlying these functions remain largely unknown. There are three lamin genes in humans. Remarkably, over 300 mutations have been identified in one of the human lamin genes, causing nearly 20 different diseases. The lamin A/C gene is one of the most highly mutated genes in humans and is unique for its association with the most diverse disease phenotypes. There is an emerging hypothesis that lamins serve a critical role in the maintenance of cell identity and integrity via regulated protein-protein interactions and associations with specific chromosome structures.The Chicago Laminome Project is a comprehensive approach to understanding lamins in health and disease via tools of systems biology. The City of Chicago had a unique set of internationally recognized leaders who have studied lamins, chromosome structure and function, and proteomics. The Chicago Laminome Project will bring this multi-institutional, multi-disciplinary group together to study the roles of lamins and lamina structure, with the longer term goal of making the Chicago area a widely-recognized center of leadership in this important field of study.






